AI predicts proteins and edits DNA
AI solves biology problems
Protein folding, a tough challenge
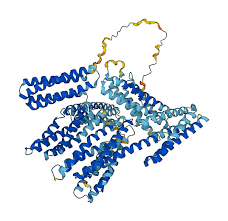
One of the longest standing-by problems of Life Sciences was protein folding.
For starters, folding is the process through which a protein is turned from a linear string of amino-acids to a three-dimensional working unit, suitable for carrying out all sorts of tasks in living cells.
This is a key problem not only because proteins are the fundamental building blocks of life, but also because misfolding (i.e. errors that occur while building the 3D structure) is at the heart of lots of human and animal diseases and abnormalities. Moreover, proteins are a target for lots of drugs (antibiotics, for example) and they can also be employed in a wide variety of industrial fields (most of clothes detergents contain enzymes to remove dirt and stains, e.g.).
In this sense, knowing the 3D structure of a protein thanks to a fast, reliable and scalable in silico method, such as a predictive algorithm, is much easier than exploiting slow and expensive laboratory procedures, such as crystallography (which is, nevertheless, still considered the golden standard for protein structure reconstruction).
For 50 years, 3D structure prediction has been a real struggle for bioinformatician, because no algorithm seemed to get accurate results, but then Google Deepmind team came, and the rules of the game changed.
Alphafold, a game changer
In 2020, AlphaFold 2 won the Critical Assessment of Structure Prediction - 14 (CASP14), a protein fold competition, with unprecedented results.
In this competition, participants are evaluated according to GDT, Global Distance Test: the score ranges from 0 to 100, with 100 meaning that all amino-acids in the predicted 3D structure of the protein are exactly where they are supposed to be, or within a threshold distance. Formally, a GDT of 90 is considered to be competitive with laboratory procedures: in 2020, AlphaFold 2 got a median score of 92.4 overall, with the lowest being around 87. Considered that no one, not even AlphaFold (base version), had surpassed 60 in the previous editions of CASP, that was a shockingly impressing result.
Now AlphaFold 2 can be used as a research tool by everyone: you can either run it within Colab (here) or you can search through the AlphaFold database, which encompasses more than 200,000,000 predicted structures: if you scrape through it long enough, you may also come across a protein structure that no one has ever seen!
So, now comes the big question: should we trust AI and rely on that for a task as delicate and critical as protein structure prediction? Share your thoughts in the comments below!
Genome editing: AI can modify human DNA
There’s a biotech startup that claims to have edited human DNA with the aid of AI: how comes that we are able to edit human DNA? How exactly AI can help in doing that? Let’s take one step at a time.
The CRISPR era
Background
Since the very first discover of DNA structure by Watson and Crick in 1953, the El Dorado of molecular biologists had been to “crack the code”: ingenerate modifications in the genetic sequence in order to change its features. The first attempts came in the 60s and 70s, were wheat plants were modified to be resistant to cold temperatures and parasites, even though these first GMO were obtained through very imprecise and coarse modifications based on some DNA absorption and/or horizontal DNA techniques already observed in bacteria (namely plasmids). Other paths that scientists used to achieve this goal were radiations, nanoparticle bombardment and viral vector transduction, but all these methods suffered from being either imprecise or highly expensive, so that no one could really be scaled on a wide-public production (one of the first medical treatment based on viral vectors for gene modification against spinal muscular atrophy, Zolgensma, costed around 2 million dollars), both for safety and financial concerns.
A revolutionary technique on the rise
Studying bacteria, scientists discovered, in 1987, that there were some intriguing repeated sequences in their DNA, which some unique and non-repetitive sequences were always found. An explanation for this characteristic came only twenty years later, when it was understood that prokaryotes use this sort of system as an “immune defense” against viruses (there is a wide class of viral agents that “eat” bacteria, known as bacteriophages).
To put it plain and simple, there was a family of proteins, the Cas family, which were able to recognize and chop the DNA of a viral invader based on previous encounters that the cell had with the same agent: in order to do so, they used a sort of “tracker”, which was actually the portion of the viral DNA they had to break. This portion was stored in the bacterial genome, and was accessible thanks to a special signal, which was actually constituted by one of the repeated sequences discovered in 1987. This system was then called CRISPR-Cas, with CRISPR meaning Clustered Regularly Interspaced Short Palindromic Repeats (luckily they found a crispy acronym for that!).
Here came the breakthrough in science: what if we implemented CRISPR-Cas system to target our genes and correct the errors that were in there? In the end, it only takes a protein (Cas9 for most of the purposes) and a guide sequence, and then the system can freely work (it is a little more complex than this, but bear with me for today’s article).
CRISPR-Cas9 was actually implemented and used for gene editing, and its impact was so massive on today’s science that in 2020, Jennifer Doudna and Emmanuelle Charpentier, two CRISPR pioneers, were awarded with the Nobel Prize in Chemistry “for the development of a method for genome editing”.
Needless to say, this comes also with several ethic implications, that even turned in a sci-fi like scenario when, in 2018, He Jiankui, a Chinese biophysicist, used CRISPR-Cas9 to modify to human embryos at make them resistant to HIV.
AI comes into the play
Regardless of ethical problems that may come along with its implementation, CRISPR-Cas9 has proved as one of the most effective and trustworthy techniques to edit DNA, and lots of experiments and clinical trials are nowadays relying on that. Nevertheless, one of the biggest problem is the unsuitability of Cas protein for the editing target: proteins are indeed molecules with an highly complex 3D structure that not always fits everywhere.
In this sense, finding alternatives to Cas9 and to the whole editing scaffold structure to generate combinations that are a perfect fit for a given situation is a key problem for scientists now. Or, better, it was a key problem: Profluent, a biotech startup, has now aced an incredible results in predicting the sequence of Cas-like proteins using generative AI models.
In their paper “Design of highly functional genome editors by modeling the universe of CRISPR-Cas sequences” they explain how they did it:
- First, they mined data from over 26 TB of assembled microbial genomes, in order to get out more than 1 million of CRISPR operons. They collected all these data in the “CRISPR-Cas9 Atlas”
- They trained an AI model (OpenCRISPR-1) in order to generate Cas9-like protein sequences
- They tested the efficacy of the editing power for their generated proteins on a human cell lines
- They collected data from other proteins involved in the editing and designed a “fully synthetic base editor system” that would best suit the outputs from OpenCRISPR-1, and tested it on the human cell lines as before.
The first results look incredible and it seems that we are less that one step away from unlocking a new frontier of gene editing, which could be used for therapeutics, fighting the climate crisis, granting access to food for everyone, optimize agricultural production, eradicate disease-bearing insects… and many other applications.
The coolest part of this project is that it is completely open source: everyone can use OpenCRISPR as long as they sign Profluent license and terms of usage to ensure ethical application of the software for research and commercial purposes.
References
- Design of highly functional genome editors by modeling the universe of CRISPR-Cas sequences Jeffrey A. Ruffolo, Stephen Nayfach, Joseph Gallagher, Aadyot Bhatnagar, Joel Beazer, Riffat Hussain, Jordan Russ, Jennifer Yip, Emily Hill, Martin Pacesa, Alexander J. Meeske, Peter Cameron, Ali Madani bioRxiv 2024.04.22.590591; doi: https://doi.org/10.1101/2024.04.22.590591
- OpenCRISPR GitHub
- A wonderful book about CRISPR: Editing Humanity: The CRISPR Revolution and the New Era of Genome Editing, by Kevin Davies (this is a spontaneous suggestion, not an advertisement)
Leave your thoughts!
What do you think about the impact of this kind of AI on health, society and environment? Is it dangerous to apply AI to genome editing?
Leave a comment below!
